Structure
Getting Start:
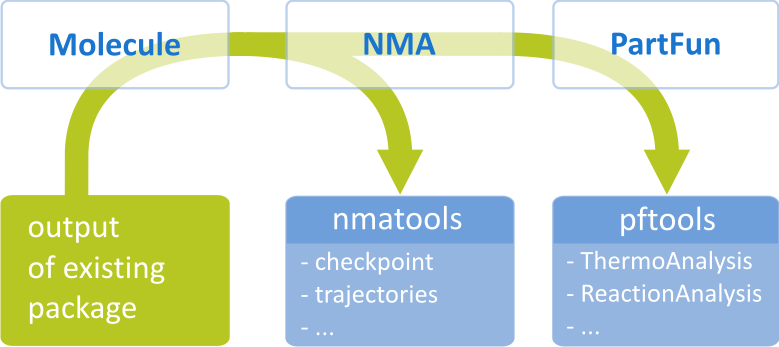
基础案例:
# Typical script #Load the TAMkin package from tamkin import * #Load the data molecule = load_molecule_g03fchk("gaussian.fchk") #Perform normal mode analysis: #If the second argument is omitted, the frequency computation is performed in 3N degrees of freedom. The ConstrainExt() variant will perform the normal mode analysis in 3N-6 (or 3N-5) internal coordinates and leads to frequencies that are identical to those of Gaussian. nma = NMA(molecule, ConstrainExt()) #Construct a partition function #A PartFun object is a definition of the partion function. All thermodynamic quantities are methods or attributes of the PartFun object. The translational and rotational contributions are included by adding [ExtTrans(), ExtRot()] as an argument. The vibrational and electronic contribution is included implicitely. pf = PartFun(nma, [ExtTrans(), ExtRot()]) # Write some general information about the molecule # and the partition function to a file. pf.write_to_file("partfun.txt") # Write an extensive overview of the thermodynamic properties to a file: ta = ThermoAnalysis(pf, [300,400,500,600]) ta.write_to_file("thermo.csv")Tamkin可以读取哪些数据:
CHARMM:输入文件需要含有
VIBRAN部分CALC k = 3 * ?NATOM VIBRan NMODes @k OPEN WRITe CARD UNIT 20 NAME filename.hessian WRITe SECOnd CARD UNIT 20 * title - Full Hessian * ENDCP2K:
The optimization:
&GLOBAL RUN_TYPE GEO_OPT PRINT_LEVEL LOW &END GLOBALThe calculation of the energy (one SCF optimization):
&GLOBAL RUN_TYPE ENERGY_FORCE PRINT_LEVEL HIGH &END GLOBALThe frequency calculation:
Gaussian 98/03/09&VIBRATIONAL_ANALYSIS FULLY_PERIODIC T &END VIBRATIONAL_ANALYSIS ... &MOTION # Optionally constrain some atoms, such that only a partial Hessian is # computed. The same constraints can be used in the optimization. # You should not use _more_ constraints in the optimization. Less is OK but # sometimes not needed. &CONSTRAINT &FIXED_ATOMS COMPONENTS_TO_FIX XYZ LIST {integer} or a range {integer}..{integer} &END FIXED_ATOMS &END CONSTRAINT &END MOTION ... &GLOBAL RUN_TYPE vibrational_analysis PRINT_LEVEL medium &END GLOBAL
Gaussian 98/03/09:
- 保持checkpoint file:
%chk=prefix.chk - 计算完成后:
formchk prefix.chk prefix.fchk
- 保持checkpoint file:
理论背景:http://molmod.github.io/tamkin/tutorial/chemphys_theory.html